Lucy student, Yi Chew, takes a One Health approach towards ending the TB epidemic by 2030.
Yi Chew is currently pursuing an MSc in Systems Biology studying pathogen genomics and evolution. Yi has always been interested in bioinformatics and molecular biology, specifically on how models can formalise and test fascinating theories and phenomena in biology. She will be joining the Bioinformatics Institute in Singapore after her graduation, and is interested in research on the dynamics between immunological interactions and pathogen evolution.
Yi submitted a blog entry about her research for the 2022 College Research Day Blog Competition, you can read it below.
Inferring interspecies transmission of Mycobacterium tuberculosis for endemic bovine TB
By Yi Chew
What is your research about?
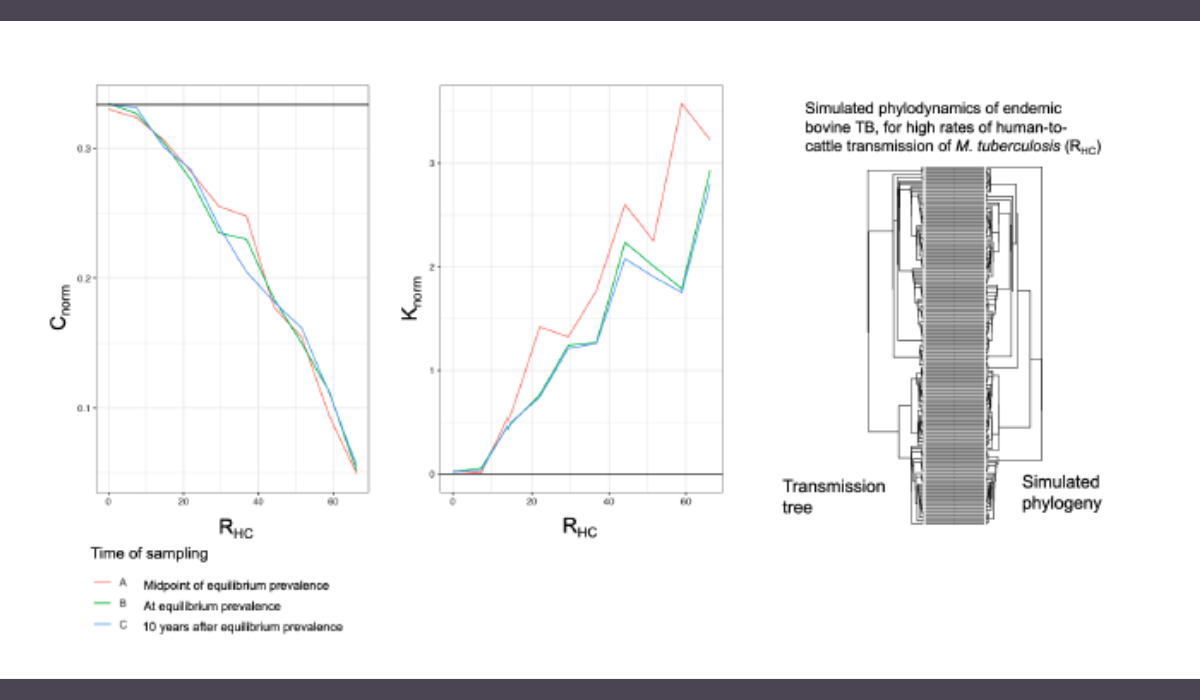
My research is in developing a framework to simulate the genomics and evolution of Mycobacterium tuberculosis in regions with endemic bovine TB and zoonotic TB in humans. This will be important for validating existing phylodynamic inference tools (i.e. TransPhylo) reconstructing transmission networks from molecular epidemiology to be used for studying endemic bovine TB. Identifying important transmission routes has implications for disease management strategies, which will be different in regions where costly test-and-slaughter strategies are incompatible with local customs and expensive to implement. In resource-poor settings, my framework for simulating phylogenies given different disease transmission scenarios will also help researchers determine the most efficient sampling strategy of cows and their human handlers for phylodynamic investigations.
Can you describe your research in one sentence?
I explore different plausible interspecies transmission scenarios for endemic bovine TB and determine if their phylogenies are sufficiently informative to be distinguished from each other, as well as the most efficient sampling strategy for obtaining informative phylogenies.
Why is this research interesting to you?
Despite having a much lower substitution rate compared to RNA viruses, for which existing phylodynamic inference tools were originally developed, M. tuberculosis phylogenies can still be informative of different underlying interspecies transmission dynamics driving the same endemic prevalence of bovine TB under ideal sampling conditions. I'm still working on determining the most effective sampling strategy to be recommended for studying endemic bovine TB.
Does your research relate to any of the UN Sustainable Development Goals?
My research takes a One Health approach towards ending the TB epidemic by 2030 under Goal 3 of the UN SDGs, by studying the contributions of both cattle and human populations towards bovine and zoonotic human TB. My phylodynamic framework can also be extended to model the impact of bovine TB vaccinations on zoonotic TB.




